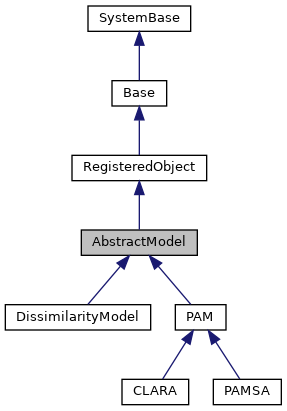
This class provides a base class for implementation for different Cluster Analysis algorithms. More...
Public Member Functions | |
| _.Library.Integer | ById (_.Library.RawString id) |
| Returns the ordinal number of the point with the given ID id. More... | |
| _.Library.Double | Distance (_.Library.Integer i, _.Library.Integer j, _.Library.Double p, _.Library.Boolean normalize) |
| Returns the dissimilarity measure between two data points of the model. More... | |
| _.Library.Double | Distance1 (_.Library.Integer i, z, _.Library.Double p, _.Library.Boolean normalize) |
| Returns the dissimilarity measure between a data points of the model and a point with given coordinates. More... | |
| _.Library.Double | Distance12 (z1, z2, _.Library.Double p, _.Library.Boolean normalize) |
| Returns the dissimilarity measure between two points with given coordinates. More... | |
| _.DeepSee.extensions.clusters.ASW | GetASWIndex () |
| Returns an object that can calculate an index used in Cluster Validation. More... | |
| _.DeepSee.extensions.clusters.CalinskiHarabasz | GetCalinskiHarabaszIndex (_.Library.Integer normalize) |
| Returns an object that can calculate an index used in Cluster Validation. More... | |
| GetCentroid (_.Library.Integer k, z) | |
| Returns the coordinates for the centroid for a given cluster. More... | |
| _.Library.Integer | GetCluster (_.Library.Integer point) |
| Returns the cluster ordinal for a given point. More... | |
| GetClusterSize (_.Library.Integer k) | |
| Returns the number of data points assigned to a given cluster. More... | |
| _.Library.Integer | GetCost (_.Library.Integer i, _.Library.Integer j) |
| Returns the dissimilarity measure as used by this clustering algorithm. More... | |
| _.Library.Integer | GetCount () |
| Returns the number of all data points in the model. | |
| _.Library.Integer | GetDimensions () |
| Returns the dimensionality of the model. | |
| _.Library.String | GetId (_.Library.Integer i) |
| Returns the unque Id of the point with the ordinal number specified by i. More... | |
| _.Library.Integer | GetNumberOfClusters () |
| Returns the number of clusters in the model. | |
| _.DeepSee.extensions.clusters.PearsonGamma | GetPearsonGammaIndex () |
| Returns an object that can calculate an index used in Cluster Validation. More... | |
| GlobalCentroid (z) | |
| Returns the coordinates for the centroid for the whole dataset. More... | |
| _.Library.Boolean | IsPrepared () |
| Checks whether the model is ready for an analysis to be executed. More... | |
| _.Library.Double | RelativeClusterCost (_.Library.Integer k, _.Library.Integer m) |
| Returns the realtive cost of a given cluster relative to a medoid point m. More... | |
| Reset () | |
| Kills all the data associated with this model. | |
| _.Library.Status | SetData (_.Library.IResultSet rs, _.Library.Integer dim, _.Library.Double nullReplacement) |
| Sets the data to be associated with this model. More... | |
| iterateCluster (_.Library.Integer k, _.Library.Integer i, _.Library.String id, coordinates) | |
| Iterates over all the data points assigned to a given cluster. More... | |
| printAll () | |
| Convenience method. More... | |
| printCluster (_.Library.Integer k) | |
| Convenience method. More... | |
 Public Member Functions inherited from RegisteredObject Public Member Functions inherited from RegisteredObject | |
| _.Library.Status | OnAddToSaveSet (_.Library.Integer depth, _.Library.Integer insert, _.Library.Integer callcount) |
| This callback method is invoked when the current object is added to the SaveSet,. More... | |
| _.Library.Status | OnClose () |
| This callback method is invoked by the <METHOD>Close</METHOD> method to. More... | |
| _.Library.Status | OnConstructClone (_.Library.RegisteredObject object, _.Library.Boolean deep, _.Library.String cloned) |
| This callback method is invoked by the <METHOD>ConstructClone</METHOD> method to. More... | |
| _.Library.Status | OnNew () |
| This callback method is invoked by the <METHOD>New</METHOD> method to. More... | |
| _.Library.Status | OnValidateObject () |
| This callback method is invoked by the <METHOD>ValidateObject</METHOD> method to. More... | |
Static Public Member Functions | |
| _.Library.Status | Delete (_.Library.String dataset) |
| Deletes a model for a dataset with the name given by dataset argument. | |
| _.Library.Boolean | Exists (_.Library.String dataset) |
| Checks whether a model for a dataset with the name given by dataset argument already exists. | |
Public Attributes | |
| DSName | |
| More... | |
| Dim | |
| More... | |
| Normalize | |
| Whether to normalize distance across multiple dimensions. More... | |
| P | |
| The power to use in calculation of dissimilarity. More... | |
| Verbose | |
| More... | |
Additional Inherited Members | |
 Static Public Attributes inherited from RegisteredObject Static Public Attributes inherited from RegisteredObject | |
| CAPTION = None | |
| Optional name used by the Form Wizard for a class when generating forms. More... | |
| JAVATYPE = None | |
| The Java type to be used when exported. | |
| PROPERTYVALIDATION = None | |
| This parameter controls the default validation behavior for the object. More... | |
This class provides a base class for implementation for different Cluster Analysis algorithms.
It defines storage for clustering models and provides methods to retrieve information about data and clustering.
Cluster analysis or clustering is the assignment of a set of observations into subsets (called clusters) so that observations in the same cluster are similar in some sense. Clustering is a method of unsupervised learning, and a common technique for statistical data analysis used in many fields, including machine learning, data mining, pattern recognition, image analysis, information retrieval, and bioinformatics.
By Default model data is stored in ^IRIS.Temp globals.
| _.Library.Integer ById | ( | _.Library.RawString | id | ) |
Returns the ordinal number of the point with the given ID id.
The unique id must correspond to the one assigned in <METHOD>SetData</METHOD>() method
| _.Library.Double Distance | ( | _.Library.Integer | i, |
| _.Library.Integer | j, | ||
| _.Library.Double | p, | ||
| _.Library.Boolean | normalize | ||
| ) |
Returns the dissimilarity measure between two data points of the model.
The method takes 4 arguments:
| _.Library.Double Distance1 | ( | _.Library.Integer | i, |
| z, | |||
| _.Library.Double | p, | ||
| _.Library.Boolean | normalize | ||
| ) |
Returns the dissimilarity measure between a data points of the model and a point with given coordinates.
The method takes 4 arguments:
| _.Library.Double Distance12 | ( | z1, | |
| z2, | |||
| _.Library.Double | p, | ||
| _.Library.Boolean | normalize | ||
| ) |
Returns the dissimilarity measure between two points with given coordinates.
The method takes 4 arguments:
| _.DeepSee.extensions.clusters.ASW GetASWIndex | ( | ) |
Returns an object that can calculate an index used in Cluster Validation.
and determining the optimal number of clusters. This method returns Average Silhouette Width index.
| _.DeepSee.extensions.clusters.CalinskiHarabasz GetCalinskiHarabaszIndex | ( | _.Library.Integer | normalize | ) |
Returns an object that can calculate an index used in Cluster Validation.
and determining the optimal number of clusters. This method returns Calinski-Harabasz index.
| GetCentroid | ( | _.Library.Integer | k, |
| z | |||
| ) |
Returns the coordinates for the centroid for a given cluster.
Cluster is identified by its ordinal number k.
Coordinates are returned as multidimensional value: z(1), z(2), ..., z(dim)
| _.Library.Integer GetCluster | ( | _.Library.Integer | point | ) |
Returns the cluster ordinal for a given point.
Point is identified by its ordinal number.
| GetClusterSize | ( | _.Library.Integer | k | ) |
Returns the number of data points assigned to a given cluster.
Cluster is identified by its ordinal number k.
| _.Library.Integer GetCost | ( | _.Library.Integer | i, |
| _.Library.Integer | j | ||
| ) |
Returns the dissimilarity measure as used by this clustering algorithm.
between two data points of the model. Points are identified by their ordinal numbers.
| _.Library.String GetId | ( | _.Library.Integer | i | ) |
Returns the unque Id of the point with the ordinal number specified by i.
The unique Id is as has been assigned in <METHOD>SetData</METHOD>() method
| _.DeepSee.extensions.clusters.PearsonGamma GetPearsonGammaIndex | ( | ) |
Returns an object that can calculate an index used in Cluster Validation.
and determining the optimal number of clusters. This method returns Pearson-Gamma index which is a correlation coefficient between distance between two points and a binary function whether they belong to the same cluster. This index is useful when clustering is used for dimension reduction i.e. the process of reducing the number of random variables under consideration
| GlobalCentroid | ( | z | ) |
Returns the coordinates for the centroid for the whole dataset.
Coordinates are returned as multidimensional value: z(1), z(2), ..., z(dim)
| _.Library.Boolean IsPrepared | ( | ) |
Checks whether the model is ready for an analysis to be executed.
This is dependent on a
specific algorithm and therefore this method is overriden by subclasses.
Reimplemented in PAM, DissimilarityModel, and CLARA.
| _.Library.Double RelativeClusterCost | ( | _.Library.Integer | k, |
| _.Library.Integer | m | ||
| ) |
Returns the realtive cost of a given cluster relative to a medoid point m.
Cluster is identified by its ordinal number k. Point m is identified by its ordinal number.
| _.Library.Status SetData | ( | _.Library.IResultSet | rs, |
| _.Library.Integer | dim, | ||
| _.Library.Double | nullReplacement | ||
| ) |
Sets the data to be associated with this model.
The method takes 3 arguments:
| iterateCluster | ( | _.Library.Integer | k, |
| _.Library.Integer | i, | ||
| _.Library.String | id, | ||
| coordinates | |||
| ) |
Iterates over all the data points assigned to a given cluster.
Cluster is identified by its ordinal number k
| printAll | ( | ) |
Convenience method.
Writes all data points in the dataset to the default output device.
| printCluster | ( | _.Library.Integer | k | ) |
Convenience method.
Writes all data points assigned to a given cluster to the default output device. Cluster is identified by its ordinal number k
| DSName |
| Dim |
| Normalize |
Whether to normalize distance across multiple dimensions.
If set to 1 (default) then
distance is normalized by variances.
| P |
The power to use in calculation of dissimilarity.
Default is Euclidean distance (P=2).
Specify 1 for Manhattan Distance or 100 for Chebyshev distance (max between coordinates).
| Verbose |